Pandas is a library for working with and manipulating tabular style data. In many ways you can think of it as a replacement for a spreadsheet only it's much more powerful. Whereas NumPy provides $N$-dimensional data structures, Pandas is best utilized on two-dimensional, labeled data. The fundamental data structures in Pandas are the Series and the Dataframe.
A Pandas Series contains a single column of data and an index. The index is a way to reference the rows of data in the Series. Common examples of an index would be simply a monotonically increasing set of integers, or time/date stamps for time series data.
A Pandas DataFrame can be thought of being created by combining more than one Series that share a common index. So a table with multiple column labels and common index would be an example of a DataFrame. The description of these data structures will be made clear through examples in the sequel.
Similarly to the way we import NumPy, it's idiomatic Python to import Pandas as
import pandas as pd
import numpy as np
import pandas as pd
Pandas offers some of the best utilities available for reading/parsing data from text files. The function read_csv has numerous options for managing header/footer lines in files, parsing dates, selecting specific columns, etc in comma separated value (CSV) files. The default index for the Dataframe is set to a set of monotonically increasing integers unless otherwise specified with the keyword argument index_col.
There are similar functions for reading Microsoft Excel spreadsheets (read_excel) and fixed width formatted text (read_fwf).
The file '200wells.csv' contains a dataset with X and Y coordinates, facies 1 and 2 (1 is sandstone and 2 interbedded sand and mudstone), porosity , permeability (mD) and acoustic impedance ($\mbox{kg} / (\mbox{m}^2 \cdot \mbox{s} \cdot 10^6)$).
The head() member function for DataFrames displays the first 5 rows of the DataFrame. Optionally, you can specify an argument (e.g. head(n=10) to display more/less rows
df = pd.read_csv('datasets/200wells.csv'); df.head()
The DataFrame member function describe provides useful summary statistics such as the total number of samples, mean, standard deviations, min/max, and quartiles for each column of the DataFrame.
df.describe()
We can access parts of the DataFrame by their labels or their numerical indices. The most basic and useful operation is to select an entire column of data by it's label.
df[['porosity']].head()
Multiple columns can be selected by passing in a list of labels.
df[['porosity', 'permeability']].head()
The member function loc can be used to select both rows and columns of data by their labels. The index is interpreted as the row label.
df.loc[1:2, ['porosity', 'permeability']]
loc support NumPy-style slicing notation to select sequences of labels.
df.loc[1:2, 'porosity':'acoustic_impedance']
The member function iloc can be used to select both rows and columns of data by their integer index. iloc supports Python-list style slicing.
df.iloc[1:3, 3:5]
We can get the underlying data in a Series/DataFrame as a NumPy array with the values attribute.
df['porosity'].values
There are several member functions that allow for transformations of the DataFrame labels, adding/removing columns, etc.
To rename DataFrame column labels, we pass a Python dictionary where the keywords are the current labels and the values are the new labels. For example,
The use of the keyword argument inplace = True has an equivalent outcome as writing
df = df.rename(...
df.rename(columns={'facies_threshold_0.3': 'facies',
'permeability': 'perm',
'acoustic_impedance': 'ai'}, inplace = True)
df.head()
Pandas DataFrames share a lot of the same syntax with Python dictionaries including accessing columns by label (i.e. keyword) and adding entries. The example below shows how to add a new column with the label 'zero'.
df['zero'] = np.zeros(len(df))
df.head()
We can remove unwanted columns with the drop member function. The argument inplace = True modifies the existing DataFrame in place in memory, i.e. 'zero' will no longer be accessible in any way in the DataFrame.
The argument axis = 1 refers to columns, the default is axis = 0 in which case the positional argument would be expected to be an index label.
df.drop('zero', axis=1, inplace=True)
df.head()
We can remove the row indexed by 1 as follows.
Notice we can stack member function commands, i.e. the drop function is immediately followed by head to display the DataFrame with row index 1 removed.
df.drop(1).head()
Because the argument inplace = True was not given, the orginal DataFrame is unchanged.
df.head()
We can sort the DataFrame in either ascending or desending order by any column label.
df.sort_values('porosity', ascending=False, inplace=True)
df.head(n=13)
In the previous example, the resulting indices are now out of order after the sorting operation. This can be fixed, if desired, with the reset_index member function.
The reindexing operation could have been accomplished during the sort operation by passing the argument ingnore_index = True.
df.reset_index(inplace=True, drop=True)
df.head(n = 13)
In the field of data science, DataFrame column labels are often referred to as features. Feature engineering is the process of creating new features and/or transforming features for further analysis. In the example below, we create two new features through manipulations of existing features.
Mathematical operations can be performed directly on the DataFrame columns that are accessed by their labels.
df['% porosity'] = df['porosity'] * 100
df['perm-por ratio'] = df['perm'] / df['porosity']
df.head()
We can also use conditional statements when assigning values to a new feature. For example, we could have a categorical porosity measure for high and low porosity, called 'porosity type'.
Most NumPy functions such as where will work directly on Pandas DataFrame columns.
df['porosity type'] = np.where(df['porosity'] > 0.12, 'high', 'low')
df[df['porosity type'] == 'high'].head(n=3)
df[df['porosity type'] == 'low'].head(n=3)
Here's an example where we use a conditional statement to assign a very low permeability value (0.0001 mD) for all porosity values below a threshold. Of course, this is for demonstration, in practice a much lower porosity threshold would likely be applied.
df['perm cutoff'] = np.where(df['porosity'] > 0.12, df['perm'], 0.0001)
df.head(n=3)
df[df['perm cutoff'] == 0.0001].head(n=3)
We can apply any function that we can define in Python to entries of a Series or DataFrame. First we define a function. This function returns the Kozeny-Carmen relationship to predict permeability from porosity ϕ, for values of ϕ greater that $0.12$, otherwise it returns $0.0001$. The proportionality constant $m$ is given as an additional keyword argument.
def kozeny_carmen_with_threshold(ϕ, m=5):
if ϕ > 0.12:
return m * ϕ ** 3 / (1 - ϕ) ** 2
else:
return 0.0001
Using the apply member function we can transform the 'porosity' column into permeability via the kozeny_carmen_with_threshold function above.
df['kozeny carmen'] = df['porosity'].apply(kozeny_carmen_with_threshold, m=10)
df
It's very common in data analysis to have missing and/or invalid values in our our DataFrames. Pandas offers several built in methods to identify and deal with missing Data.
First, let's create a missing data point in the 'porosity' column of df.
The at member function allows for fast selection of a single row/label position within a Series/DataFrame.
df.at[1, 'porosity'] = None
df.head(n = 3)
The isnull member function returns a boolean array that identifies rows with missing values.
df[df['porosity'].isnull()]
The dropna member function allows us to remove missing rows from the DataFrame.
df.dropna(inplace=True)
df.head(n = 3)
Up to now, we've utilized a DataFrame that was created by reading in the data from a CSV file. We can also create DataFrames from scratch. First we'll create a couple of NumPy arrays with representative data for demonstration.
porosity = np.random.random_sample(100) * 0.2
permeability = 10 * porosity ** 3 / (1 - porosity) ** 2
Create a Python dictionary where the keywords are the desired DataFrame labels and the values are the associated data.
df_dict = {'porosity': porosity, 'permeability': permeability}
Pass the dictionary as an argument to pd.Dataframe to create the DataFrame
df_new = pd.DataFrame(df_dict)
df_new.sort_values('porosity', inplace=True, ignore_index=True)
df_new.head()
In this example, we'll take a couple subsets from our original DataFrame and create a new one by joining them with the concat function.
df1 = df.iloc[0:3,2:]; df1
df2 = df.iloc[10:13,2:]; df2
We could have reindexed with the ingnore_index = True keyword argument.
pd.concat([df1, df2], axis=0)
Pandas has some built in automatic plotting methods for DataFrames. They are most useful for quick-look plots of relationships between DataFrame columns, but they can be fully customized to make publication quality plots with additional options available in the Matplotlib plotting library. The default plot member function will create a line plot of all DataFrame labels as functions of the index.
df_new.plot();
Of course, this is not that useful a plot. What we are likely looking for is a relationship between the DataFrame columns. One way to visualize this is to set the desired abscissa as the DataFrame index and create a plot.
df_new.set_index(['porosity']).plot();
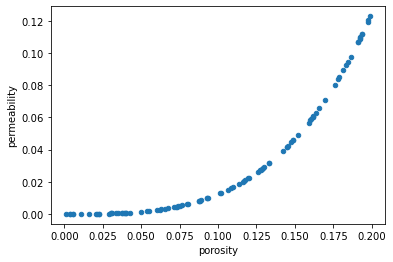
Or explicitly pass the desired independent variable to the x keyword arguments and the dependent variable to the y keyword argument. This time we'll also explicitly create a scatter plot.
When the DataFrame columns are explicitly passed as arguments, the axis labels are correctly populated.
df_new.plot.scatter(x='porosity', y='permeability');
The most common way way to store and share DataFrames among different computers is the export the DataFrame to a CSV file. This is accomplished with the to_csv member function.
df.to_csv("datasets/200wells_out.csv", index = False)
Displaying the raw data is the file with the UNIX head command
!head -n 5 datasets/200wells_out.csv
Further Reading
Further reading on Pandas is avialable in the official Pandas documentation